Design and Application Guidelines for RNA Primers in PCR Technology
Integrating Technical Expertise, Structural Insights, and Advanced Methodologies
I. Applications and Advantages of RNA Primers
1. Key Use Cases
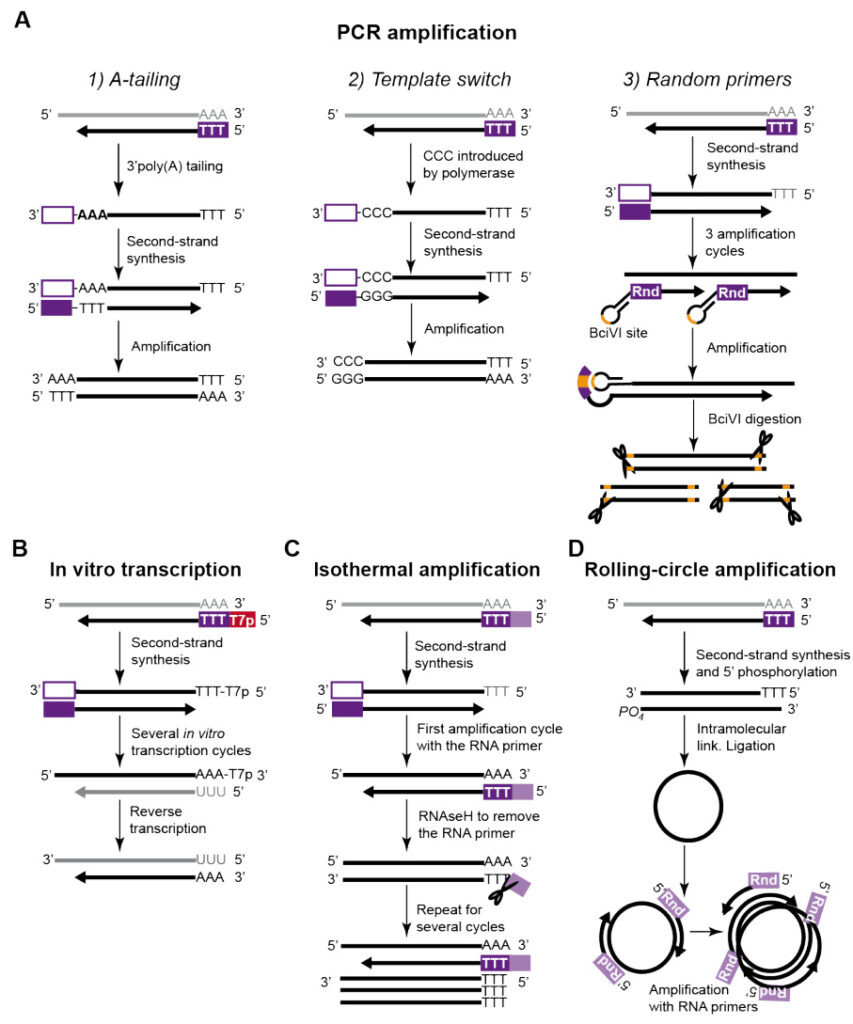
- Reverse Transcription PCR (RT-PCR): RNA primers bind to poly-A tails or specific sequences (e.g., viral RNA genomes) to initiate DNA synthesis from RNA templates.
- RNA-Primed PCR: Utilizes reverse transcriptase-active DNA polymerases (e.g., rTth polymerase) for direct DNA amplification without prior cDNA synthesis.
- Single-Cell RNA Sequencing (scRNA-seq): Employs random RNA primers (e.g., N6 primers) for whole-transcriptome amplification, minimizing template loss.
2. Core Advantages
- Cost Efficiency: Bulk synthesis via in vitro transcription reduces costs compared to chemically synthesized DNA primers.
- Dynamic Control: Thermolabile modifications (e.g., 2′-O-methyl) enable temperature-dependent primer activation, enhancing amplification specificity.
II. Critical Parameters for RNA Primer Design
1. Sequence Design Principles
- Length: Typically 4–12 nucleotides (nt) for rapid binding; extended to 30–35 nt for specialized applications (e.g., T7 promoter primers).
- GC Content: Maintain 40–60% to minimize secondary structures; avoid consecutive G/C repeats.
- 3′-End Stability: Terminate with G/C bases to enhance elongation efficiency.
2. Structural Optimization Strategies
- Chemical Modifications:
- 2′-O-Methylation: Protects against RNase degradation during high-temperature cycling.
- Phosphorothioate Bonds: Stabilize primer-template complexes.
- Functional Elements:
- T7 Promoter Sequences: Facilitate downstream in vitro transcription.
- Molecular Barcodes: Enable multiplexed amplification (e.g., scRNA-seq).
3. Specificity Controls
- Exon-Intron Spanning: Design primers across exon-exon junctions to avoid genomic DNA contamination.
- Bioinformatics Validation: Use tools like Primer-BLAST (NCBI) or RNAstructure to predict secondary structures and off-target binding.
III. Experimental Workflow and Optimization
1. Enzyme and Reaction Setup
- Reverse Transcriptase Selection: Use thermostable enzymes (e.g., rTth polymerase) compatible with RNA primers.
- Manganese Ion Optimization: Adjust Mn²⁺ concentration (1–3 mM) to balance reverse transcription and DNA polymerization.
2. Thermal Cycling Adjustments
- Denaturation Temperature: Reduce to 85–90°C to minimize RNA primer degradation.
- Extension Time: Extend to 2–5 minutes to accommodate lower elongation efficiency of RNA primers.
3. Contamination Mitigation
- RNase Inhibitors: Add RNasin (1 U/μL) to reaction buffers.
- Physical Separation: Prepare RNA primers in a dedicated workspace to avoid cross-contamination.
IV. Innovative Technologies and Emerging Applications
1. CRISPR-Guided Primer Systems
- Cas13-dgRNA Complexes: Target RNA primers to specific regions for isothermal amplification (e.g., Recombinase Polymerase Amplification, RPA).
2. Spatiotemporally Controlled Priming
- Photoactivatable Primers: Incorporate photocleavable groups (e.g., PC-biotin) for light-triggered activation in live cells.
3. Multiplex Amplification Strategies
- Nested RNA Primers: Improve sensitivity for low-abundance targets via two-round amplification.
V. Troubleshooting Common Issues
| Issue | Root Cause | Solution |
|---|---|---|
| Nonspecific Amplification | Primer secondary structures or off-target binding | Redesign sequences or add stabilizing modifications |
| Low Amplification Efficiency | Primer degradation or insufficient polymerase activity | Optimize Mn²⁺ levels or increase cycle number (up to 50 cycles) |
| Long-Fragment Detection Failure | Limited primer elongation capacity | Switch to chimeric RNA-DNA primers (e.g., 5′-DNA-modified primers) |
VI. Future Perspectives
RNA primers are pivotal in specialized PCR applications (e.g., viral RNA detection, single-cell genomics), but their design requires balancing enzyme compatibility, structural stability, and experimental conditions. Emerging trends include:
- AI-Driven Design Tools: Predict RNA primer-template interaction dynamics for enhanced specificity.
- Multifunctional Chimeric Primers: Integrate CRISPR targeting with signaling modules (e.g., molecular beacons).
- Microfluidic Integration: Enable high-throughput synthesis and screening of RNA primers.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




