 1. Molecular Foundations of CRISPR Targeting
1. Molecular Foundations of CRISPR Targeting
CRISPR systems rely on precise interactions between Cas endonucleases (e.g., Cas9, Cas12a) and guide RNAs (gRNAs) to recognize and cleave target DNA sequences. The gRNA, comprising a CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) or a single-guide RNA (sgRNA), directs Cas proteins to complementary genomic loci via Watson-Crick base pairing . Target recognition requires two critical elements:
- Protospacer Adjacent Motif (PAM): A short DNA sequence (e.g., 5′-NGG-3′ for S. pyogenes Cas9) adjacent to the target site, essential for Cas protein activation .
- Seed Sequence: A 10–12 nt region at the 3′ end of the gRNA spacer that initiates DNA hybridization .
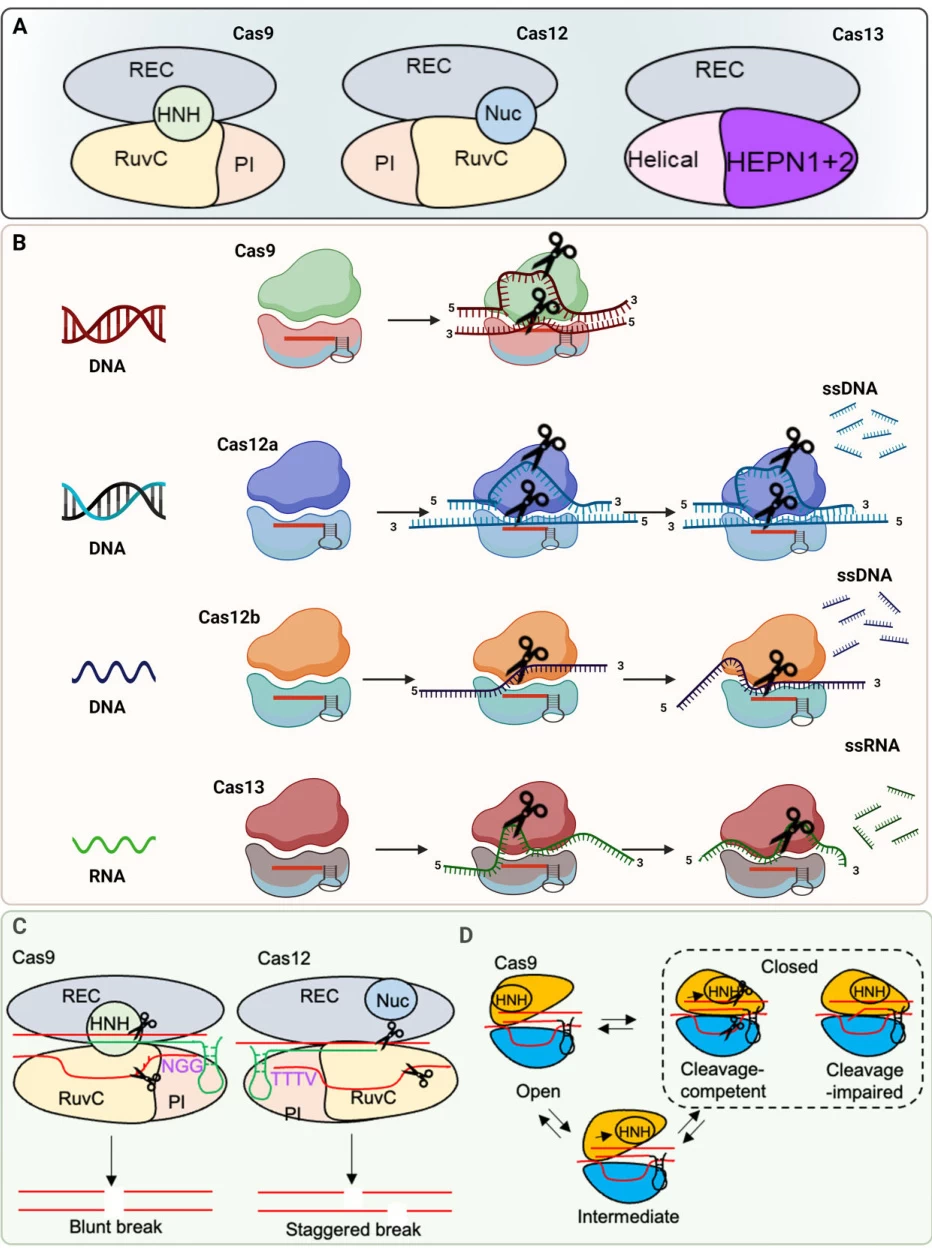
Structural Determinants:
- Cas9: Forms a bilobed structure upon sgRNA binding. The PAM-interacting (PI) domain recognizes PAM, triggering DNA unwinding and gRNA-DNA hybridization. The HNH domain cleaves the complementary strand, while the RuvC-like domain cleaves the non-complementary strand, generating blunt-ended double-strand breaks (DSBs) .
- Cas12a: Recognizes a 5′-TTTV-3′ PAM upstream of the target. Its single RuvC domain cleaves both DNA strands, producing staggered DSBs .
2. gRNA Design Principles for Optimal Targeting
A. Sequence-Specific Optimization
- Complementarity:
- A 20-nt spacer provides ideal balance between specificity and efficiency. Mismatches in the PAM-proximal “seed” region (positions 1–12) disrupt cleavage more severely than distal mismatches .
- GC Content:
- Maintain 40–60% GC content to prevent secondary structures that impair Cas binding .
- Avoidance of Problematic Motifs:
- Exclude poly-T stretches (≥4 T) to prevent transcriptional termination and repetitive sequences to minimize off-target effects .
B. Conformational Dynamics
Cas9 exists in three states:
- Open State: sgRNA-loaded but inactive.
- Intermediate State: Partial DNA hybridization.
- Closed State: Fully hybridized, catalytically active conformation .
Design Implication: gRNAs with high on-target affinity accelerate transition to the closed state.
3. Enhancing Efficiency Through Protein Engineering
A. High-Fidelity Cas Variants
| Variant | Key Mutation | Efficiency Gain |
|---|---|---|
| eSpCas9 | K848A/K1003A/F1085A | 10–100× specificity |
| Cas9-HF1 | N497A/R661A/Q695A/Q926A | >90% on-target |
| HypaCas9 | N692A/M694A/Q695A/H698A | 5× fewer off-targets |
B. Cas12a Advantages
- Simpler gRNA: Requires only crRNA (no tracrRNA).
- Collateral Activity: Cleaves non-target ssDNA after activation, enabling ultrasensitive diagnostics .
4. Computational & Experimental Validation Workflow
Key Validation Methods:
- T7 Endonuclease I (T7E1) Assay: Detects indels at target sites.
- GUIDE-seq: Genome-wide mapping of off-target cleavages .
- Single-Molecule FRET: Quantifies real-time Cas9-DNA binding kinetics .
5. Advanced Applications & Case Studies
A. Diagnostic Systems
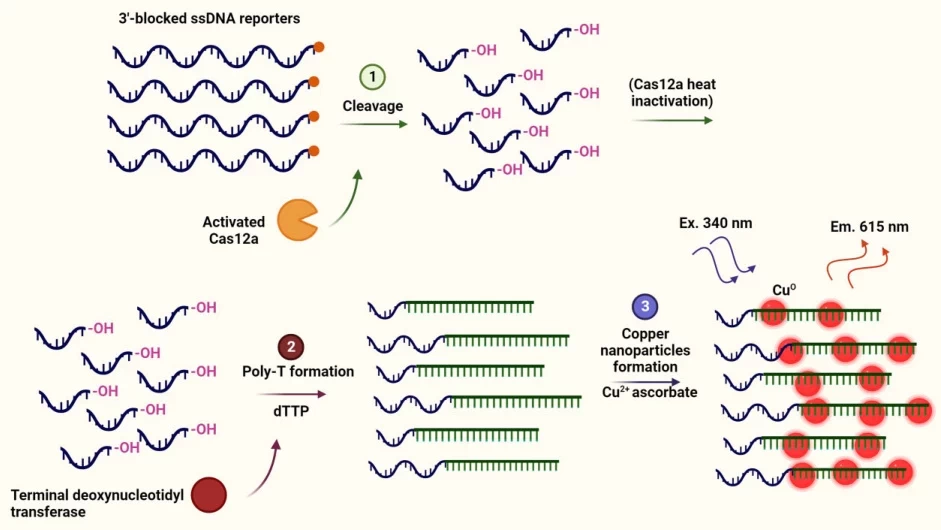
- CANTRIP (Cas12a-activated Nanoparticle Reporter):
- Cas12a cleaves target DNA.
- Collateral ssDNA cleavage generates 3′-OH ends.
- Terminal transferase adds poly-T tails.
- Copper nanoparticles form, emitting fluorescence for detection .
Sensitivity: Detects attomolar SARS-CoV-2 RNA.
B. Therapeutic Genome Editing
- CRISPR-Cas9 for HIV-1:
- gRNAs target HIV proviral DNA integrated into host genomes.
- Cas9 cleavage triggers error-prone NHEJ, disrupting viral genes .
- Base Editing:
- Fusion of nickase Cas9 (nCas9) with deaminases enables C→T or A→G conversions without DSBs .
6. Emerging Frontiers
- Cell-Type-Specific Targeting:
- Integrate scATAC-seq data to design gRNAs for lineage-specific enhancers .
- Quantum Computing Optimization:
- Predict Cas9-gRNA binding affinity using quantum annealing algorithms .
- In Vivo Delivery Systems:
- Lipid nanoparticles encapsulating chemically modified gRNAs (e.g., 2′-O-methyl) enhance stability .
Conclusion
Maximizing CRISPR-target efficiency requires synergistic optimization of:
- gRNA Design: 20-nt spacers, 40–60% GC, and PAM-proximal complementarity.
- Cas Protein Selection: High-fidelity variants or Cas12a for diagnostics.
- Context Awareness: Chromatin accessibility and cellular environment.
Advances in machine learning and structural biology will enable de novo design of CRISPR systems for previously “undruggable” genomic targets.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




