Architectural Precision and Functional Complexity in Life’s Blueprint
I. Chromosomes as Information Storage Systems
Chromosomes are meticulously organized DNA-protein complexes that safeguard genetic instructions critical for organismal development, function, and evolution. Their structural and functional hierarchy ensures both stability and accessibility of genetic data.
1. Hierarchical DNA Packaging
- Nucleosomes: DNA wraps around histone octamers (H2A, H2B, H3, H4) to form nucleosomes—the fundamental units of chromatin .
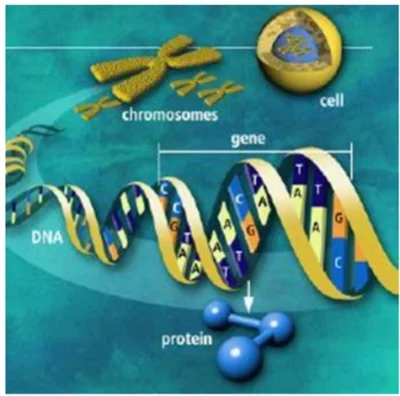
- 30-nm Fiber: Nucleosomes coil into a solenoid structure stabilized by histone H1, enabling efficient spatial compaction .
- Chromosome Scaffolding: Non-histone proteins (e.g., cohesins, condensins) organize chromatin into looped domains anchored to a protein scaffold, achieving 10,000-fold compaction .
Image suggestion: Multiscale visualization of DNA packaging from nucleosomes to metaphase chromosomes.
2. Genetic Code Organization
- Genes: Functional DNA segments (~1-2% of human DNA) encode proteins or regulatory RNAs. A single chromosome may carry thousands of genes .
- Non-Coding Regions: Satellite DNA (highly repetitive sequences) ensures chromosome stability, while regulatory elements (promoters, enhancers) control gene expression .
II. Mechanisms of Genetic Information Transmission
1. DNA Replication: Fidelity in Duplication
- Semi-Conservative Replication: Each DNA strand serves as a template for synthesizing complementary strands, preserving sequence accuracy .
- Replisome Machinery: DNA helicase unwinds the double helix, while DNA polymerases (α, δ, ε in eukaryotes) extend primers with proofreading exonuclease activity .
Image suggestion: 3D animation of replication forks showing leading/lagging strand synthesis and Okazaki fragment ligation.
2. Mitosis: Equitable Distribution
- Prophase to Telophase: Chromosomes condense, align at the metaphase plate, and segregate into daughter cells via microtubule-driven forces .
- Checkpoint Controls: The spindle assembly checkpoint (SAC) prevents aneuploidy by delaying division until all kinetochores attach correctly .
3. Meiosis: Diversity Through Recombination
- Homologous Pairing: Synaptonemal complexes mediate alignment and crossover between homologs, shuffling alleles .
- Chiasma Formation: Site-specific DNA breaks (SPO11-induced) are repaired via homologous recombination, generating genetic diversity .
Image suggestion: Cross-sectional view of crossover events during prophase I, highlighting chiasmata and recombinant chromatids.
III. Chromosomal Safeguards for Genetic Integrity
1. Telomeres: Guardians of Chromosome Ends
- Structure: TTAGGG repeats bound by shelterin proteins (TRF1, TRF2) prevent end-to-end fusion and exonuclease degradation .
- Telomerase: In germ and stem cells, this ribonucleoprotein extends telomeres, counteracting replication-associated shortening .
2. DNA Repair Systems
- Base Excision Repair (BER): Corrects small base lesions (e.g., deamination) via glycosylases and DNA polymerase β .
- Homologous Recombination (HR): Error-free repair of double-strand breaks using sister chromatid templates during S/G2 phases .
Image suggestion: Schematic comparing BER, NHEJ, and HR pathways with key enzyme annotations.
IV. Chromosomes in Gene Expression Regulation
1. Epigenetic Modifications
- Histone Acetylation: Acetyltransferases (e.g., p300) loosen chromatin, facilitating transcription factor access .
- DNA Methylation: Methylation at CpG islands (e.g., gene promoters) recruits repressive proteins (MeCP2), silencing genes .
2. 3D Genome Architecture
- Topologically Associating Domains (TADs): Insulated neighborhoods coordinate tissue-specific gene activation/silencing .
- Chromosome Territories: Non-random nuclear positioning influences transcriptional activity (e.g., gene-rich chromosomes occupy central regions) .
V. Chromosomal Aberrations and Human Health
1. Aneuploidy
- Trisomy 21 (Down Syndrome): Extra chromosome 21 disrupts developmental gene networks, causing cognitive and physical abnormalities .
- Cancer: Chromosomal instability (CIN) drives tumor evolution by amplifying oncogenes (e.g., MYC) or deleting tumor suppressors (e.g., TP53) .
2. Structural Rearrangements
- Philadelphia Chromosome: t(9;22) translocation creates the BCR-ABL1 fusion oncogene in chronic myeloid leukemia .
- Copy Number Variations (CNVs): Duplications/deletions (>1 kb) contribute to autism, schizophrenia, and rare genetic disorders .
Image suggestion: Karyotype highlighting common aneuploidies and translocations with clinical annotations.
VI. Evolutionary and Technological Implications
1. Chromosomes in Speciation
- Robertsonian Fusions: Centric fusions (e.g., human chromosome 2) reduce diploid numbers, contributing to reproductive isolation .
- Neofunctionalization: Duplicated genes acquire novel roles (e.g., olfactory receptor diversification in mammals) .
2. CRISPR-Based Engineering
- Chromosome Editing: Prime editing enables precise modifications (e.g., correcting sickle cell mutations in HBB) .
- Synthetic Chromosomes: Designer chromosomes with customized gene clusters advance synthetic biology and bioremediation .
Conclusion
Chromosomes exemplify nature’s solution to balancing genetic stability with adaptive flexibility. From their nanoscale DNA packaging to macroscale roles in heredity and disease, chromosomes remain central to understanding life’s molecular logic. Emerging technologies—from single-cell epigenomics to chromosome-scale assembly—promise to unlock new frontiers in medicine, agriculture, and synthetic life.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




