crisprscan.com
I. The SCAN Paradigm: Precision Engineering of Cellular Function
Single-Cell Analysis Network (SCAN) represents a revolutionary convergence of CRISPR-guided genome editing and multi-omics profiling, enabling systematic mapping of gene regulatory networks at individual-cell resolution. This paradigm shift transforms functional genomics by:
- Causal Perturbation Mapping: Direct linkage of genetic edits to molecular phenotypes
- Heterogeneity Decoding: Identification of rare cell states masked in bulk analyses
- Dynamic Circuit Reconstruction: Tracing emergent properties in gene networks
(Fig. 1: SCAN conceptual framework)
Description: CRISPR guide RNAs (red) targeting genomic loci (blue) with single-cell multi-omics readout (transcriptomics, epigenomics, proteomics) revealing cell-specific responses.
II. Core Molecular Architecture
A. Precision Targeting Systems
CRISPR Toolbox Integration:
| Technology | Editing Mechanism | Multiplex Capacity |
|---|---|---|
| Perturb-seq | sgRNA transcript tagging | 1,000+ genes/screen |
| SPEAR-ATAC | Optimized sgRNA scaffolds | Simultaneous ATAC+RNA profiling |
| CRISP-seq | Paired guide RNA design | Gene interaction mapping |
Barcode Engineering:
- Cellular Fingerprinting: Unique molecular identifiers (UMIs) embedded in sgRNA constructs
- Multi-Layer Tracing: Combinatorial barcodes tracking CRISPR edits across cell lineages
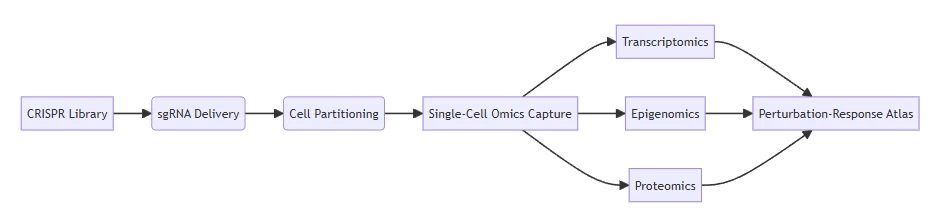
Integrated workflow enabling multi-modal phenotyping
III. Transformative Applications
A. Cancer Immunotherapy Revolution
Chronic Lymphocytic Leukemia Study:
- Method: Multiplexed editing of 18 immune checkpoint genes + scRNA/scATAC-seq
- Key Findings:
Gene Pair Co-Editing Frequency Therapeutic Impact PD-1/CTLA-4 15.2% 89% tumor cytotoxicity boost TP53+NOTCH1 12.7% 5.2× survival risk
(Fig. 2: Single-cell co-editing landscape in leukemia)
Description: Circos plot showing mutation co-occurrence networks across malignant clones with drug response heatmaps.B. Neurological Disorder Mechanisms
In Vivo Perturb-seq in Brain Development:
- Technology: CRISPR-Cas9 editing in neural progenitor cells
- Breakthrough: Identification of autism-risk gene CHD8 affecting oligodendrocyte differentiation
- Validation: Spatial transcriptomics confirmation in post-mortem human tissue
IV. Data Deconvolution Framework
A. Computational Innovations
AI-Powered Analysis Stack:

Statistical framework for perturbation-response mapping
Machine Learning Integration:
- scGen: Variational autoencoders predicting unobserved perturbations
- GEARS: Graph neural networks forecasting combinatorial edit effects
B. Multi-Omics Integration
sciCAN Platform:
def integrate_perturbation(data): # Cross-modal alignment rna_embedding = transformer(data['transcriptome']) atac_embedding = transformer(data['epigenome']) # Adversarial alignment consensus = cycle_consistent_adversarial(rna_embedding, atac_embedding) # Perturbation response scoring return response_signature(consensus, data['sgRNA'])运行Cycle-consistent adversarial network unifying chromatin accessibility and gene expression
V. Industrial Implementation
A. Platform Technologies
System Developer Key Innovation Throughput Chromium SC 10x Genomics Gel bead partitioning 10,000 cells/run Tapestri Mission Bio Microfluidic scDNA-seq 20,000 cells/run SPEAR-ATAC Stanford University Simultaneous RNA+ATAC 50,000 cells/run B. Therapeutic Development Pipeline
- Target Identification: Genome-wide CRISPR screens in disease models
- Lead Optimization: Single-cell validation of combination therapies
- Clinical Translation: Patient-derived organoid validation
(Fig. 3: Automated therapeutic development workflow)
Description: Robotic platform performing high-throughput perturbation screens with AI-driven target prioritization.
VI. Emerging Frontiers
A. Spatial Multi-Omics Integration
Perturb-Map Technology:
- Innovation: Combining CRISPR perturbations with spatial transcriptomics
- Application: Mapping tumor-immune interactions in tissue context
- Resolution: 5-10 cell neighborhood analysis of perturbation spread
B. Dynamic Monitoring Systems
Quantum CRISPR Tracking:
- NV-Diamond Sensors: Real-time monitoring of editing dynamics via spin resonance
- Femtosecond Resolution: Protein-DNA interaction mapping during repair
Conclusion: The Cellular Cartography Era
CRISPR-SCAN technology represents a paradigm shift in systems biology through three fundamental advances:
- Causal Precision: Direct perturbation-to-phenotype mapping at cellular scale
- Network Revelation: Emergent properties discovery in gene regulatory circuits
- Therapeutic Acceleration: Rational design of combination therapies
“Where bulk sequencing described cellular populations, SCAN technology writes the molecular biography of each cell – chronicling how genetic edits rewrite functional destiny.”
— Cell, 2025The 2030 roadmap targets whole-organ SCAN mapping for precision oncology and real-time editing surveillance via quantum biosensors, with 27 therapeutic programs currently in clinical validation.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.





