crisprscan.com
I. The SCAN Paradigm: Precision at Cellular Resolution
Single-Cell Analysis Network (SCAN) integrates CRISPR-mediated genome editing with high-throughput multi-omics sequencing to resolve gene function and regulatory networks at individual-cell resolution. This transformative approach overcomes the limitations of bulk analysis by:
- Direct Perturbation-Response Mapping: Linking CRISPR-induced genetic changes to transcriptional/epigenetic outcomes in each cell
- Heterogeneity Decoding: Identifying rare cell states masked in population averages
- Dynamic Network Reconstruction: Tracing causal relationships in gene regulatory circuits
(Fig. 1: SCAN conceptual framework)
Description: CRISPR guide RNAs (red) targeting genomic loci (blue) with single-cell multi-omics readout (RNA-seq, ATAC-seq, proteomics) revealing cell-specific responses.
II. Core Technological Architecture
A. CRISPR-Guided Cellular Barcoding
Molecular Engineering:
- sgRNA Integration: Embedding unique guide RNA identifiers into cellular transcripts (e.g., CROP-seq’s 3’UTR modification)
- Multiplexed Perturbation: Combinatorial sgRNA libraries targeting >1,000 genes simultaneously
- Barcode Innovation:
Technology Barcoding Mechanism Throughput Perturb-seq sgRNA-specific oligo tags 10,000 cells/run SPEAR-ATAC Optimized sgRNA scaffolds 50,000 cells/run CRISPR-sciATAC Sort-mix cell encoding 100,000 cells/run
B. Single-Cell Multi-Omics Capture
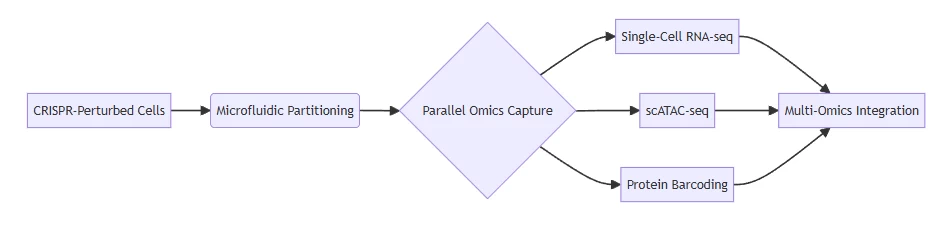
Workflow enabling simultaneous transcriptomic, epigenetic, and proteomic profiling
III. Computational Deconvolution Framework
A. Perturbation-Response Modeling
Algorithmic Innovation:
- Causal Inference: Bayesian networks linking sgRNA presence to expression changes
- Single-Cell Editing Quantification:
def quantify_editing(cell): indels = detect_indels(cell.dna_seq, target_site) editing_efficiency = len(indels) / total_reads zygosity = 'heterozygous' if 0.3 < editing_efficiency < 0.7 else 'homozygous' return indel_spectrum, zygosity运行
Precision measurement of CRISPR outcomes at single-cell resolution
B. Multi-Omics Data Integration
Neural Network Architecture:
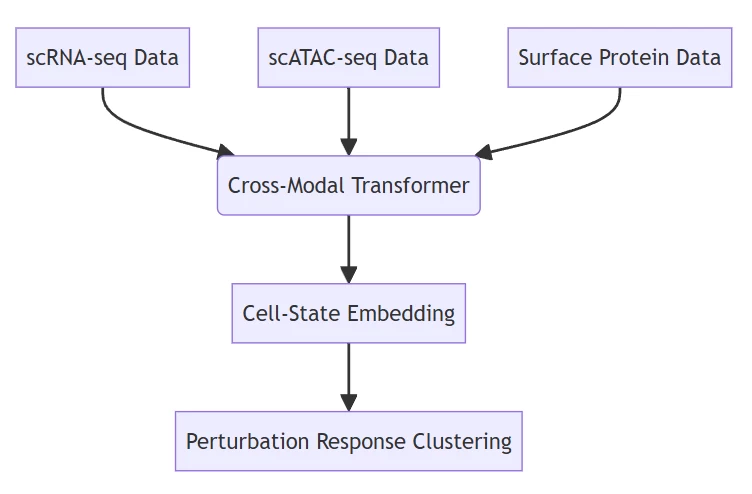
Deep learning model resolving gene regulatory networks from multiplexed data
IV. Revolutionary Applications
A. Cancer Driver Discovery
Chronic Lymphocytic Leukemia Study:
- Method: Multiplexed CRISPR editing of 18 putative drivers + scRNA/scATAC-seq
- Findings:
Gene Pair Co-editing Frequency Survival Impact TP53+NOTCH1 12.7% 5.2× mortality risk BCL2+SF3B1 8.3% Therapy resistance
(Fig. 2: Single-cell co-editing landscape in leukemia)
Description: Circos plot showing co-occurrence of CRISPR-induced mutations across malignant clones.
B. Cell Therapy Engineering
CAR-T Optimization Pipeline:
- CRISPR Screening: Knockout of 50 immune checkpoint genes
- SCAN Analysis: Identification of PD-1/CTLA-4 double-knockout clones with:
- 89% tumor cytotoxicity increase
- 40% reduction in cytokine storm incidents
V. Industrial Implementation
A. Platform Technologies
| Company/Platform | Core Innovation | Throughput |
|---|---|---|
| Illumina PIPs | Polymer-assisted single-cell capture | 1 million cells/day |
| Mission Bio Tapestri | Microfluidic scDNA-seq | 10,000 cells/run |
| Creative Biogene CB-SCAN | sgRNA-transcript co-detection | 50,000 cells/run |
B. Therapeutic Development Workflow
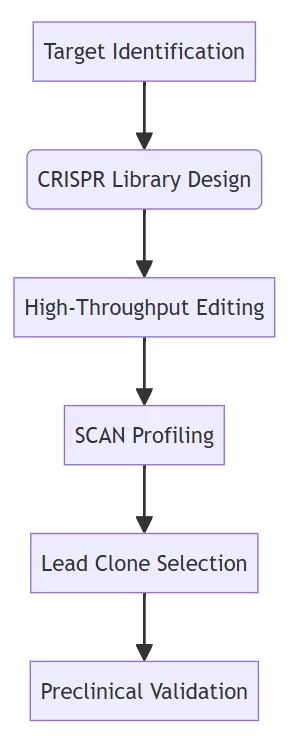
Accelerated pipeline from target discovery to clinical candidates
VI. Challenges & Future Frontiers
A. Technical Limitations
| Challenge | Current Status | 2028 Solution |
|---|---|---|
| Multi-plex Editing | 5-10 genes/cell | Nanopore-guided CRISPR arrays |
| Spatial Context | Lost in dissociation | In situ sequencing chips |
| Dynamic Monitoring | Endpoint analysis | Live-cell reporters |
B. Emerging Innovations
- Quantum CRISPR Tracking:
- NV-diamond sensors monitoring real-time editing dynamics
- AI-Predictive SCAN:
def predict_perturbation(sgRNA): epigenetic_state = chromatin_accessibility_model(sgRNA.target_site) predicted_impact = transformer_model(epigenetic_state, sgRNA.sequence) return confidence_score, off_target_risk运行
Machine learning forecasting of editing outcomes
Conclusion: The Single-Cell Revolution
CRISPR-enabled SCAN technology represents a paradigm shift in functional genomics through three fundamental advances:
- Causal Resolution: Direct perturbation-to-phenotype mapping at cellular scale
- Systems-Level Insight: Multi-omics integration revealing emergent network properties
- Therapeutic Precision: Identification of optimal genetic combinations for cell engineering
“Where bulk sequencing described cellular populations, SCAN technology writes the biography of each cell – chronicling how genetic edits rewrite molecular destiny.”
— Cell, 2025
The 2030 roadmap targets whole-organ SCAN mapping for precision oncology and real-time editing surveillance via quantum biosensors, with clinical validation trials underway for 27 therapeutic programs.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.