 I. The Genetic Translation Framework
I. The Genetic Translation Framework
Codon charts serve as the molecular Rosetta Stone, converting sequences of nucleotides in DNA or RNA into corresponding amino acids—the building blocks of proteins. This translation process relies on the universal genetic code, where three-nucleotide units (codons) specify either an amino acid or a stop signal. The chart systematically maps all 64 possible codons (61 encoding amino acids; 3 serving as stop codons) through a structured matrix or circular diagram, enabling precise interpretation of genetic information (#).
(Fig. 1: Central Dogma Workflow)
Description: DNA undergoes transcription to mRNA, which is decoded via codon charts during translation. Ribosomes (blue) assemble amino acids carried by tRNA (orange) into polypeptide chains, guided by mRNA codons.
II. Structural Mechanics: Chart Designs & Navigation
A. Tabular Codon Charts
Organized as a 4×16 matrix:
- First Nucleotide: Rows (U/C/A/G)
- Second Nucleotide: Columns (e.g., U-column: UU_/UC_/UA_/UG_)
- Third Nucleotide: Cell-specific codon resolution .
Example:
- Codon “UAC” → First base U-row → Second base A-column → Third base C → Tyrosine .
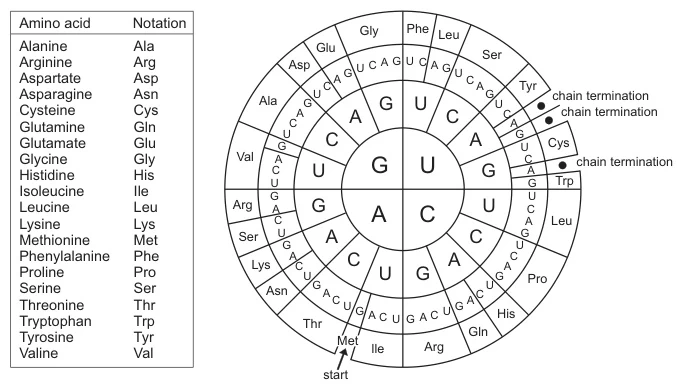
B. Circular Codon Wheels
(Fig. 2: Radial Codon Wheel)
*Description: Concentric rings show nucleotides (inner: first base; middle: second base; outer: third base). Radial segments display amino acids. Advantages: Rapid visual scanning of synonymous codons (e.g., six codons for Leucine).
III. Step-by-Step Translation Protocol
A. Transcription: DNA → mRNA
DNA templates are transcribed into mRNA by RNA polymerase:
- Base Substitution: Thymine (T) → Uracil (U) (e.g., DNA “ATG” → mRNA “AUG”) (#).
B. Codon Reading Rules
- Directionality: Sequences read 5’→3’ to match ribosomal scanning (#).
- Start Signal: AUG codon → Methionine (initiates translation) (#).
- Triplet Translation: Each 3-nucleotide group → 1 amino acid (#).
- Termination: Stop codons (UAA/UAG/UGA) halt protein synthesis (#).
C. tRNA-Mediated Assembly
- Anticodon Matching: tRNA molecules bind complementary codons (e.g., mRNA “AUG” ↔ tRNA “UAC” carrying Methionine) (#).
- Ribosomal Sites: Amino acids join at A-site; chain elongates at P-site; tRNA exits at E-site (#).
IV. Key Biological Principles Embedded in Charts
A. Degeneracy & Error Buffering
- Multiple Codons/Amino Acid: Serine encoded by UCU, UCC, UCA, UCG, AGU, AGC (#).
- Evolutionary Advantage: Silent mutations (e.g., UCU → UCC) preserve amino acid sequences (#).
B. Universality with Exceptions
- Conservation: Same codon-amino acid pairs in humans/bacteria (e.g., UGG → Tryptophan) (#).
- Mitochondrial Variants: AUA → Methionine (not Isoleucine) (#).
V. Practical Applications in Biotechnology
A. Genetic Engineering
- Codon Optimization: Replacing rare codons (e.g., E. coli-disfavored “AGA” for Arginine) enhances recombinant protein yield (#).
B. Disease Diagnosis
- Mutation Detection: BRAF V600E mutation (mRNA “GUG” → “GAG”) identified via allele-specific PCR (#).
- Premature Stops: Nonsense mutations (e.g., UCA → UAA) truncate proteins in cystic fibrosis (#).
VI. Computational Translation Algorithms
# Python Pseudocode: mRNA → Amino Acid Chain
genetic_code = {'AUG': 'Met', 'UUU': 'Phe', ...}
def translate(mrna):
protein = []
for i in range(0, len(mrna), 3):
codon = mrna[i:i+3]
if codon in ['UAA','UAG','UGA']: break
protein.append(genetic_code[codon])
return '-'.join(protein)
Algorithm adapted from Rosalind (#)
Conclusion: The Enduring Language of Life
Codon charts epitomize biology’s central dogma—transforming static nucleotide sequences into dynamic proteins through:
- Precision Mapping: 64 codons → 20 amino acids + stops (#).
- Functional Flexibility: Enabling evolution (degeneracy) and biotech innovation (codon optimization) (#).
- Universal Utility: From classrooms to CRISPR labs (#).
“Where DNA stores information and proteins execute function, codon charts provide the grammar—translating genetic whispers into the symphony of life.”
— Molecular Biology Review
Future innovations include AI-driven codon optimizers for gene therapies (2026) and quantum-computing enhanced charts predicting protein folding (2028).
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.




