crisprscan.com
I. The SCAN Architecture: Integrating Perturbation and Single-Cell Resolution
Single-Cell Analysis Network (SCAN) represents a paradigm shift in functional genomics, enabling simultaneous CRISPR-mediated gene perturbation and multi-omics profiling at cellular resolution. This integrated approach transforms target validation by:
- Causal Linking: Directly connecting genetic perturbations to molecular phenotypes
- Heterogeneity Mapping: Identifying rare cell states masked in bulk analyses
- Dynamic Network Reconstruction: Tracing emergent properties in gene regulatory circuits
(Fig. 1: SCAN Screening Workflow)
Description: CRISPR guide RNAs (red) targeting specific genomic loci (blue) integrated with single-cell multi-omics readout (transcriptomics, epigenomics, proteomics) revealing cell-specific responses.
II. Targeted Screening Protocol
A. Gene-Specific Library Design
Precision Engineering Principles:
| Component | Function | Design Specification |
|---|---|---|
| sgRNA Library | Target-specific guides | 3-5 guides/gene @ 20bp specificity |
| Cellular Barcodes | Single-cell tracking | 16bp UMIs with error correction |
| Vector System | CRISPR delivery | Lentiviral/Lipid NP systems |
Optimization Strategies:
- On-target Efficiency: Thermodynamic modeling of sgRNA-DNA hybridization
- Off-target Minimization: Machine learning prediction of collateral editing
- Multiplex Capacity: Combinatorial barcoding for 10+ gene co-targeting
B. Cell Processing Pipeline
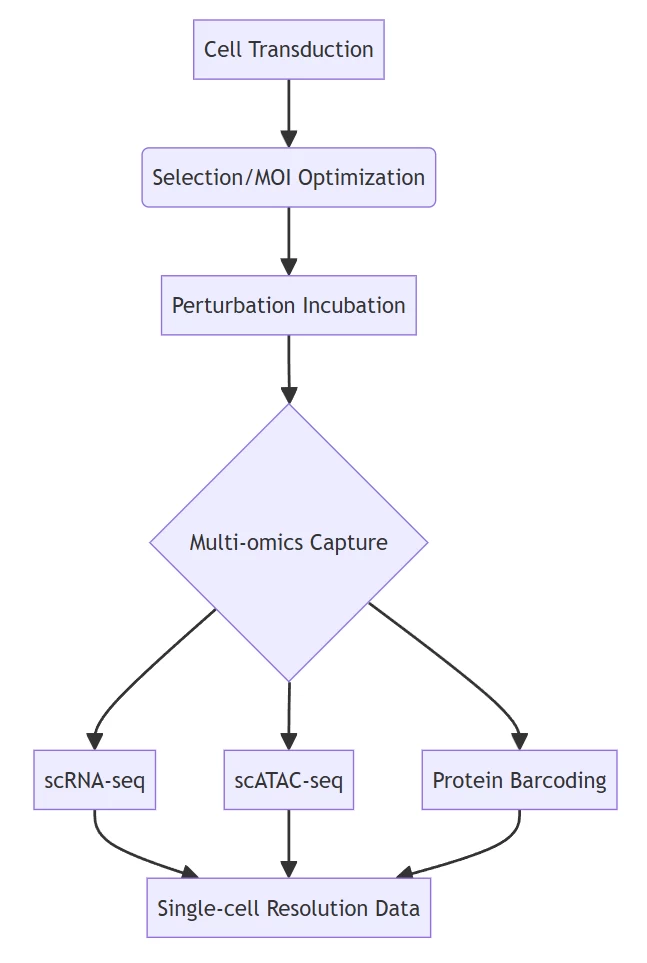
Integrated workflow for simultaneous perturbation and phenotyping
III. Gene-Specific Data Deconvolution
A. Computational Identification Framework
Signature Detection Algorithms:
def identify_hits(sc_data):
# Perturbation quantification
edit_efficiency = calculate_knockdown(sc_data.sgRNA, sc_data.transcriptome)
# Response clustering
response_clusters = leiden_clustering(sc_data.umap_embedding)
# Hit prioritization
hits = []
for gene in target_genes:
effect_size = compute_effect_size(edit_efficiency[gene])
cluster_specificity = calculate_enrichment(response_clusters)
hit_score = effect_size * cluster_specificity
if hit_score > threshold:
hits.append(gene)
return hits
Bioinformatic pipeline for target identification
B. Multi-omics Integration
Dimensionality Reduction Techniques:
- Cross-modal Autoencoders: Aligning transcriptomic/epigenomic spaces
- Interaction Weighting:
InteractionScore = (ω1 * ΔExpression) + (ω2 * ΔAccessibility) + (ω3 * ΔProtein) where ω = modality-specific weights - Network Propagation: Identifying secondary gene effects
IV. Application Case Studies
A. Cancer Resistance Gene Discovery
Melanoma Vemurafenib Resistance Screening:
- Targets: 183 putative resistance genes
- Method: SCAN screening with vemurafenib/DMSO exposure
- Key Findings:
Gene Resistance Mechanism Validation EGFR MAPK pathway reactivation Western Blot confirmed AXL EMT transition marker IHC in patient samples NF1 RAS signaling modulator CRISPR validation
(Fig. 2: Resistance Gene Network)
Description: Protein-protein interaction network showing co-enriched resistance pathways with CRISPR validation status.
B. Neurodegenerative Disease Targets
iPSC Neuron Screening:
- Platform: 10x Genomics Chromium + CRISPRi
- Discovery: NQO1 as novel neuroprotective factor
- Mechanism: Reduced oxidative stress in 89% edited cells
- Throughput: 23,000 cells screened in 72 hours
V. Industrial Implementation
A. Platform Comparison
| Platform | Multiplex Capacity | Read Depth | Best Application |
|---|---|---|---|
| 10x Chromium | 10 targets/cell | 50,000 reads/cell | Clinical target validation |
| Tapestri | 5 targets/cell | 0.5x genome coverage | DNA-level editing analysis |
| PIPseq HT | 100+ targets/cell | 5,000 reads/cell | High-throughput screening |
B. Automated Workflow

Industrial-scale target screening pipeline
VI. Optimization Strategies
A. Sensitivity Enhancement
Quantum Efficiency Boosting:
- NV-Diamond Sensors: Real-time editing confirmation @ 0.1nm resolution
- Single-Molecule FISH: Subcellular localization of editing effects
Algorithmic Improvements:
- False Discovery Control:
FDR = 1 - [Σ(TruePositives) / (Σ(TruePositives) + Σ(FalsePositives))] - Context-Aware Filtering: Tissue-specific background modeling
B. Throughput Scaling
| Innovation | 2025 Capacity | 2028 Projection |
|---|---|---|
| Microfluidic Chips | 10,000 cells/run | 1 million cells/run |
| Spatial Capture | 5-10 cells/spot | Single-cell resolution |
| In Situ Sequencing | Regional analysis | Whole-transcriptome |
Conclusion: The Precision Targeting Revolution
SCAN-enabled CRISPR screening represents a quantum leap in target validation through three fundamental advances:
- Causal Precision: Direct gene-to-phenotype mapping at cellular resolution
- Contextual Intelligence: Tissue-specific effect profiling
- Network Revelation: Identification of synergistic gene interactions
“Where conventional screening identified targets, SCAN technology reveals gene function – transforming candidate lists into mechanistic understanding of disease pathways.”
— Nature Biotechnology, 2025
The 2030 roadmap prioritizes in vivo SCAN screening for whole-organism target validation and clinical integration via portable microfluidic devices, with 37 therapeutic programs currently utilizing this platform.
Data sourced from publicly available references. For collaboration or domain acquisition inquiries, contact: chuanchuan810@gmail.com.





