1. Fundamental Definitions
sgRNA (Single Guide RNA)
A synthetic RNA molecule central to the CRISPR-Cas9 genome editing system. It combines the functions of two natural RNAs—crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA)—into a single chimeric molecule. sgRNA directs the Cas9 nuclease to specific DNA sequences for precise gene editing, enabling knockouts, knock-ins, or base modifications.
siRNA (Small Interfering RNA)
A short (~20–25 nucleotide) double-stranded RNA molecule involved in RNA interference (RNAi). siRNA binds to the RNA-induced silencing complex (RISC), guiding it to complementary mRNA targets for degradation or translational repression, thereby silencing gene expression post-transcriptionally.
2. Structural and Functional Contrasts
A. Molecular Architecture
| Feature | sgRNA | siRNA |
|---|---|---|
| Composition | Single-stranded RNA (~100–200 nt) with a crRNA-tracrRNA fusion | Double-stranded RNA (~20–25 bp) with a guide strand and passenger strand |
| Key Domains | – Protospacer (20 nt DNA-targeting sequence) – tracrRNA scaffold (stem-loops for Cas9 binding) |
– Guide strand (complementary to mRNA) – Passenger strand (discarded) |
| Chemical Modifications | 2′-O-methylation, phosphorothioate bonds for stability in mammalian cells | Chemical modifications (e.g., 2′-O-methyl) to enhance RISC loading and reduce immune activation |
Image suggestion: Side-by-side structural models of sgRNA (showing crRNA-tracrRNA fusion) and siRNA (highlighting duplex structure and RISC binding).
B. Mechanism of Action
| Process | sgRNA | siRNA |
|---|---|---|
| Target Recognition | Binds DNA via Watson-Crick pairing near a protospacer adjacent motif (PAM) | Binds mRNA via perfect complementarity to the guide strand |
| Enzyme Recruitment | Directs Cas9 to induce double-strand DNA breaks (DSBs) | Recruits RISC to cleave or repress mRNA |
| Outcome | Permanent genomic edits (knockouts, knock-ins) | Transient gene silencing (mRNA degradation or translational inhibition) |
Image suggestion: Mechanistic flowchart comparing sgRNA-Cas9 DNA cleavage vs. siRNA-RISC mRNA degradation.
3. Design and Optimization Strategies
A. sgRNA Design
- Target Selection: Requires a 20-nt protospacer adjacent to a PAM (e.g., 5′-NGG-3′ for S. pyogenes Cas9). Tools like CRISPR-ERA predict on-target efficiency and off-risk scores.
- Scaffold Engineering: Modified stem-loop structures (e.g., MS2 aptamers) enable recruitment of transcriptional activators/repressors (CRISPRa/i).
B. siRNA Design
- Seed Region Optimization: Nucleotides 2–8 of the guide strand determine specificity; mismatches here reduce off-target effects.
- Chemical Modifications: 2′-O-methyl or locked nucleic acids (LNAs) enhance stability and reduce immunogenicity.
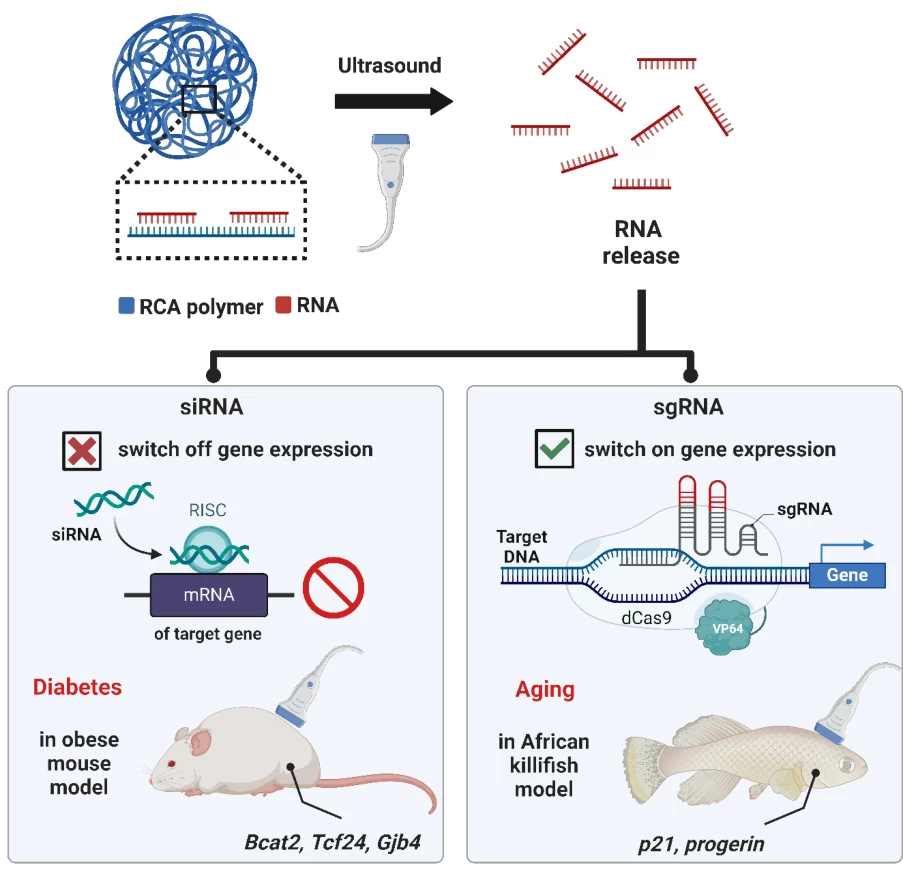
4. Applications in Research and Therapy
| Application | sgRNA | siRNA |
|---|---|---|
| Gene Knockout | Permanent disruption via NHEJ or HDR (e.g., GFP knockout in HeLa cells) | Not applicable (targets mRNA, not DNA) |
| Gene Silencing | Indirect via dCas9-sgRNA fusions (e.g., CRISPRi) | Direct mRNA degradation (e.g., Bcat2 silencing in obesity models) |
| Therapeutic Use | In vivo editing for genetic disorders (e.g., sickle cell anemia) | Treatment of viral infections (e.g., hepatitis C) or cancers |
Image suggestion: Comparative table with icons representing applications (e.g., DNA scissors for sgRNA, mRNA breakdown for siRNA).
5. Challenges and Solutions
A. Off-Target Effects
- sgRNA: Mismatches in the seed region (nt 1–12) can cause unintended DNA edits. Solutions include high-fidelity Cas9 variants (e.g., HypaCas9).
- siRNA: Off-target mRNA binding due to partial complementarity. Mitigated via seed region optimization and bioinformatics screening.
B. Delivery Systems
- sgRNA: Delivered via lipid nanoparticles (LNPs) or viral vectors (e.g., AAV).
- siRNA: LNPs or GalNAc conjugates for liver-targeted delivery (e.g., patisiran for amyloidosis).
6. Emerging Innovations
- sgRNA: Prime editing (PE) systems enable precise base changes without DSBs.
- siRNA: Self-delivering RNA (sdRNA) with chemical modifications for enhanced cellular uptake.
Conclusion
sgRNA and siRNA represent two pillars of modern molecular biology—genome editing and gene silencing, respectively. While sgRNA enables permanent DNA modifications through CRISPR-Cas9, siRNA offers transient mRNA silencing via RNAi. Their structural, mechanistic, and functional distinctions define unique roles in research and therapy. Advances in design tools (e.g., CRISPR-ERA), delivery systems (e.g., LNPs), and precision engineering (e.g., prime editing) continue to expand their transformative potential.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




