The Molecular Compass of CRISPR-Cas9 Gene Editing
I. Fundamental Definition
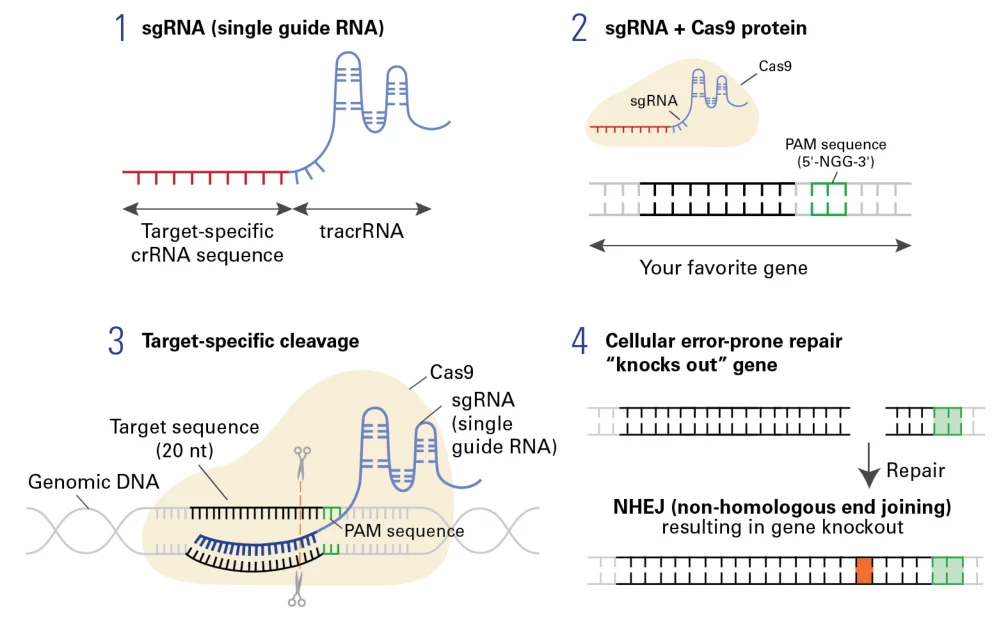
Small guide RNA (sgRNA) is a synthetic, single-stranded RNA molecule that serves as the targeting module in the CRISPR-Cas9 genome editing system. It combines two natural RNA components—crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA)—into a single chimeric molecule, enabling precise guidance of the Cas9 nuclease to specific DNA sequences .
Key Characteristics:
- Length: Typically 100–200 nucleotides (nt), with a 20-nt protospacer sequence for DNA targeting .
- Function: Directs Cas9 to induce double-strand breaks (DSBs) at predefined genomic loci, enabling knockouts, knock-ins, or base editing .
II. Structural Architecture
sgRNA exhibits a modular design optimized for both target recognition and Cas9 binding:
1. Target-Specific Region (crRNA)
- Protospacer: A 20-nt sequence complementary to the target DNA adjacent to a protospacer adjacent motif (PAM) (e.g., 5′-NGG-3′ for Streptococcus pyogenes Cas9) .
- Role: Ensures sequence-specific DNA hybridization via Watson-Crick base pairing .
2. Cas9-Binding Scaffold (tracrRNA)
- Stem-Loop Structures: Conserved hairpin motifs (e.g., nexus, stem-loop 1–3) that bind Cas9’s REC lobe, stabilizing the ribonucleoprotein (RNP) complex .
- Chemical Modifications: 2′-O-methyl or phosphorothioate bonds enhance stability and reduce immune recognition in mammalian cells .
Image suggestion: Ribbon diagram of sgRNA-Cas9 complex highlighting protospacer-PAM interaction and tracrRNA scaffolding.
III. Mechanism of Action
The sgRNA-Cas9 complex operates through a three-step process:
1. Target Recognition
- PAM Scanning: Cas9 slides along DNA, identifying PAM motifs through base-specific interactions .
- DNA Hybridization: The protospacer pairs with the complementary DNA strand, triggering Cas9 conformational changes .
2. DNA Cleavage
- Nuclease Domains: Cas9’s HNH and RuvC-like domains cleave the target and non-target DNA strands, respectively, generating DSBs .
3. Repair and Editing
- Non-Homologous End Joining (NHEJ): Error-prone repair often introduces insertions/deletions (indels), disrupting gene function .
- Homology-Directed Repair (HDR): Template-dependent repair enables precise edits in the presence of donor DNA .
Image suggestion: Stepwise illustration of sgRNA-guided DNA cleavage and repair pathways.
IV. Design Principles for Optimal Efficiency
Effective sgRNA design balances specificity, on-target activity, and minimal off-target effects:
1. Target Selection
- Exon Prioritization: For gene knockouts, target early exons to maximize frameshift mutations .
- PAM Proximity: Position protospacer 3–8 nt upstream of functional domains (e.g., catalytic sites) for precise editing .
2. Sequence Optimization
- GC Content: Maintain 30–70% to avoid secondary structures and ensure stable hybridization .
- Off-Target Screening: Use tools like CRISPOR or Cas-OFFinder to predict and exclude sequences with ≥4 mismatches .
3. Delivery Format
- Synthetic sgRNA: Chemically modified RNAs (e.g., Alt-R™) enhance stability and editing efficiency .
- Expression Vectors: U6 or T7 promoters drive sgRNA transcription in plasmids or viral vectors .
Image suggestion: Flowchart of sgRNA design workflow with bioinformatics validation steps.
V. Applications in Research and Therapy
1. Functional Genomics
- Genome-Wide Screens: Libraries of sgRNAs (e.g., Brunello) enable high-throughput identification of genes regulating traits like drug resistance .
- Cell-Specific Editing: Tissue-specific promoters or logic-gated sgRNAs restrict Cas9 activity to defined cell types .
2. Therapeutic Development
- In Vivo Editing: Lipid nanoparticle (LNP)-encapsulated sgRNA/Cas9 treats genetic disorders (e.g., sickle cell anemia) .
- Cancer Immunotherapy: sgRNAs targeting PD-1 enhance T-cell antitumor activity .
VI. Challenges and Innovations
1. Off-Target Effects
- Mismatch Tolerance: Even 1–2 mismatches in seed regions (nt 1–12) can cause unintended edits .
- Solutions: High-fidelity Cas9 variants (e.g., HypaCas9) or paired nickases reduce off-target activity .
2. Delivery Bottlenecks
- Size Limitations: Viral vectors (e.g., AAV) struggle to package Cas9-sgRNA cassettes >4.7 kb .
- Innovations: Split-Cas9 systems or RNA virus vectors circumvent size constraints .
Conclusion
sgRNA has revolutionized genome engineering by merging simplicity with precision. Its dual role as a molecular GPS and nuclease activator underpins breakthroughs from basic research to clinical trials. Continued advances in chemical biology, computational design, and delivery technologies promise to unlock sgRNA’s full potential in curing genetic diseases and redefining biotechnology.
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




