 Exploring the Diversity and Functional Roles of Translation Termination Signals
Exploring the Diversity and Functional Roles of Translation Termination Signals
1. Introduction to Stop Codons
Stop codons are nucleotide triplets within messenger RNA (mRNA) that signal the termination of protein synthesis during translation. These codons do not encode amino acids but instead recruit release factors (RFs) to dissociate ribosomal subunits and release nascent polypeptide chains. Their role as “stop signs” in genetic translation ensures the accurate production of functional proteins .
2. Standard Stop Codons in the Universal Genetic Code
The canonical genetic code recognizes three stop codons:
- UAA (Ochre): The most universally conserved termination signal across species.
- UAG (Amber): First identified in bacteriophage studies, named after researcher Harris Bernstein (“Bernstein” means “amber” in German).
- UGA (Opal or Umber): Dual-functional in some contexts, encoding selenocysteine in eukaryotes when accompanied by a SECIS element (selenocysteine insertion sequence) .
DNA Equivalents: During transcription, DNA sequences TAA, TAG, and TGA are converted into the RNA stop codons UAA, UAG, and UGA, respectively .
Image suggestion: Ribosomal complex model showing stop codon (UGA) recognition by release factor eRF1, with mRNA and nascent polypeptide chain.
3. Non-Canonical and Species-Specific Variants
While the three codons above dominate most organisms, exceptions exist in specialized genetic systems:
A. Mitochondrial Stop Codons
- Vertebrate Mitochondria: Use AGA and AGG as additional stop codons, repurposing these arginine codons in nuclear DNA.
- Green Algae (Scenedesmus obliquus): Reassign UGA to encode tryptophan, requiring alternative termination signals like UCA .
B. Ciliate Protozoa and Genetic Code Reassignment
- Euplotes and Parduczia: All three standard stop codons (UAA, UAG, UGA) encode amino acids—glutamine or cysteine—depending on mRNA context. Translation termination relies on poly(A) tail proximity and ribosomal positioning rather than codon identity .
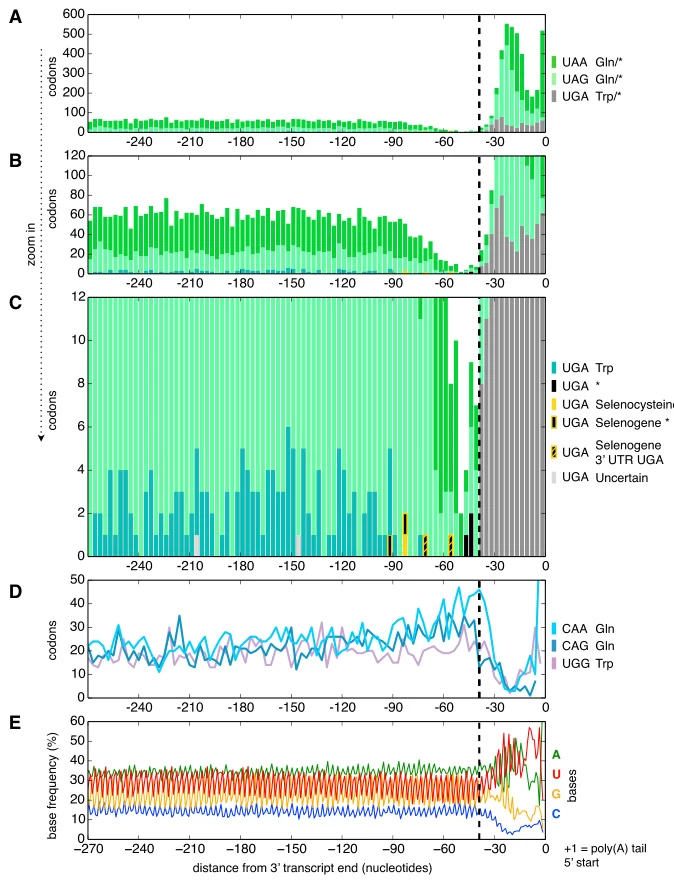
- Alternative Termination Mechanisms: In Condylostoma magnum, stop codon usage is restricted to transcript termini, minimizing premature termination risks .
Image suggestion: Phylogenetic tree highlighting stop codon reassignment in ciliates, mitochondria, and algae.
4. Stop Codon Recognition and Termination Machinery
A. Release Factors
- Prokaryotes:
- RF1: Recognizes UAA and UAG.
- RF2: Recognizes UAA and UGA.
- RF3: GTPase aiding ribosomal subunit dissociation.
- Eukaryotes:
- eRF1: Universal factor recognizing all three stop codons. Structural studies reveal its tRNA-like domain interacting with the ribosomal decoding center .
B. Structural Insights
Cryo-EM studies show that stop codons adopt a stacked conformation in the ribosomal A-site, positioning the +4 nucleotide (immediately downstream) to stabilize eRF1 binding. This “tetranucleotide motif” ensures termination fidelity .
Image suggestion: Cryo-EM structure of eRF1 bound to UAA, highlighting base stacking and ribosomal RNA interactions.
5. Biological Implications and Applications
A. Nonsense Mutations and Disease
Premature stop codons (PTCs) caused by point mutations truncate proteins, leading to diseases like cystic fibrosis (CFTR mutations) and Duchenne muscular dystrophy (DMD mutations). PTCs account for ~11% of inherited disorders .
B. Readthrough Therapies
Compounds like ataluren promote ribosomal readthrough of PTCs, restoring partial protein function. Clinical trials show promise for nonsense-mediated cystic fibrosis and muscular dystrophy .
C. Synthetic Biology
- Genome Editing: CRISPR-Cas9 introduces stop codons to knock out genes (e.g., PD-1 in CAR-T cells).
- Expanded Genetic Codes: Engineered tRNAs suppress stop codons to incorporate noncanonical amino acids, enabling novel protein functionalities .
Image suggestion: Workflow of CRISPR-Cas9 introducing a stop codon to disrupt PD-1 in T cells.
6. Evolutionary Perspectives
Stop codons likely originated from ambiguous primordial codons co-opted for termination. Their redundancy (three codons for one function) buffers against mutations, while reassignment in ciliates and mitochondria exemplifies genetic code plasticity .
Summary Table: Stop Codon Classification and Properties
| Codon (RNA) | Name | DNA Template | Species-Specific Reassignment | Key Features |
|---|---|---|---|---|
| UAA | Ochre | TAA | None (universal) | Most conserved, high termination efficiency |
| UAG | Amber | TAG | Glutamine (ciliates) | First discovered in phage studies |
| UGA | Opal | TGA | Selenocysteine (eukaryotes), Tryptophan (mitochondria) | Dual coding in SECIS-containing mRNAs |
| AGA | – | AGA | Stop (vertebrate mitochondria) | Repurposed arginine codon |
| AGG | – | AGG | Stop (vertebrate mitochondria) | Repurposed arginine codon |
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




