 Precision, Mechanism, and Evolutionary Significance in Genetic Translation
Precision, Mechanism, and Evolutionary Significance in Genetic Translation
1. Core Definition and Molecular Architecture
A stop codon (or termination codon) is a triplet of nucleotides within messenger RNA (mRNA) that signals the termination of protein synthesis during translation. These codons do not encode amino acids but instead recruit release factors (RFs) to dissociate ribosomal subunits and release the nascent polypeptide chain.
- RNA Sequences: UAA (“ochre”), UAG (“amber”), and UGA (“opal”) .
- DNA Equivalents: TAA, TAG, and TGA, transcribed into RNA during mRNA synthesis.
- Functional Analogy: Acts as a “stop sign” in translation, analogous to a period concluding a sentence.
Image suggestion: Molecular model of a ribosome bound to mRNA, highlighting the stop codon (UGA) in the A-site and the release factor (eRF1) interaction.
2. Mechanism of Translation Termination
Stop codons are recognized by release factors, proteins that catalyze the hydrolysis of the peptidyl-tRNA bond, freeing the polypeptide chain.
A. Prokaryotic Systems
- RF1: Recognizes UAA and UAG.
- RF2: Recognizes UAA and UGA.
- RF3: GTPase that assists in ribosome dissociation.
B. Eukaryotic Systems
- eRF1: A universal release factor recognizing all three stop codons. Structural studies reveal that eRF1 interacts not only with the stop codon but also with the +4 base (the nucleotide immediately downstream), forming a “tetranucleotide motif” critical for termination fidelity .
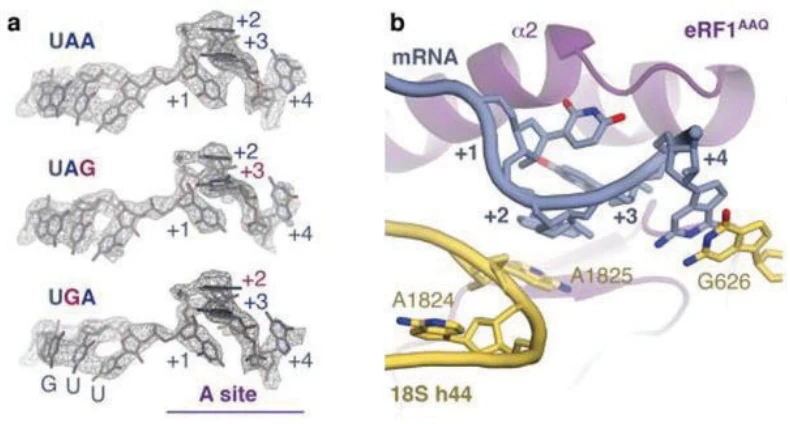
- Conformational Dynamics: Stop codons in the ribosomal A-site adopt a stacked conformation, pulling the +4 base into proximity with decoding center residues (e.g., G626 in eukaryotes), enhancing termination accuracy .
Image suggestion: Cryo-EM structure of eRF1 bound to a stop codon (UAA) and the ribosomal decoding center, illustrating base stacking and conformational changes.
3. Evolutionary and Biological Significance
A. Genetic Code Redundancy
The existence of three stop codons provides redundancy, reducing the risk of translational errors due to single-nucleotide mutations. For example:
- UGA Dual Function: In certain contexts, UGA encodes selenocysteine, a rare amino acid critical for antioxidant enzymes .
- Mitochondrial Variations: Some organisms (e.g., humans) utilize expanded stop codons (AGA, AGG) in mitochondrial translation .
B. Species-Specific Adaptations
- Bacteria and Archaea: Preferential use of UAA over UAG/UGA reflects evolutionary pressure for termination efficiency.
- Viruses: Some viral genomes minimize stop codons to force ribosomal readthrough, enabling synthesis of longer polyproteins .
Image suggestion: Phylogenetic tree showing stop codon usage preferences across species, with annotations for selenocysteine and mitochondrial variants.
4. Mutational Consequences and Disease
A. Nonsense Mutations
Premature stop codons caused by point mutations (e.g., CAG→UAG) result in truncated, nonfunctional proteins. Examples include:
- Cystic Fibrosis: CFTR nonsense mutations (e.g., G542X) disrupt chloride channel function .
- Duchenne Muscular Dystrophy: DMD truncations lead to loss of dystrophin, causing muscle degeneration .
B. Readthrough Therapies
Small molecules (e.g., ataluren) promote ribosomal readthrough of premature stop codons, restoring partial protein function. Clinical trials show promise for diseases like nonsense-mediated cystic fibrosis .
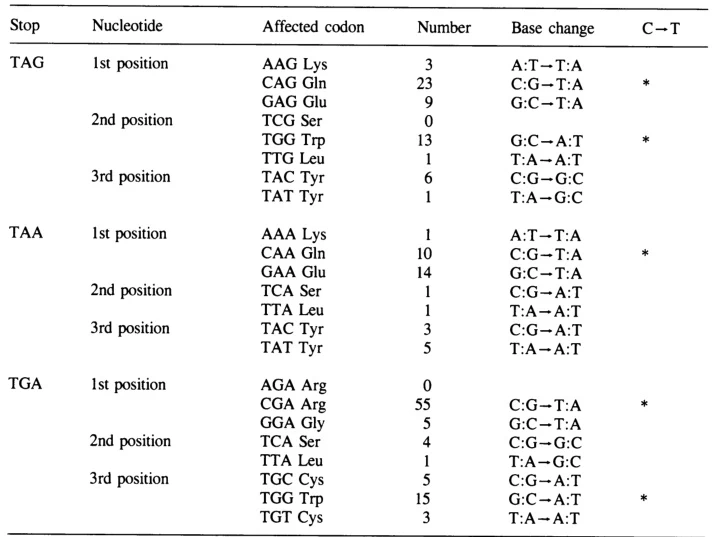
Image suggestion: Mechanistic diagram of readthrough therapy: Drug-induced bypass of a premature stop codon (UAG) in the CFTR mRNA.
5. Applications in Biotechnology and Synthetic Biology
A. Gene Knockout and Engineering
- CRISPR-Cas9: Introduction of stop codons disrupts target genes. For example, truncating PD-1 in CAR-T cells enhances antitumor activity .
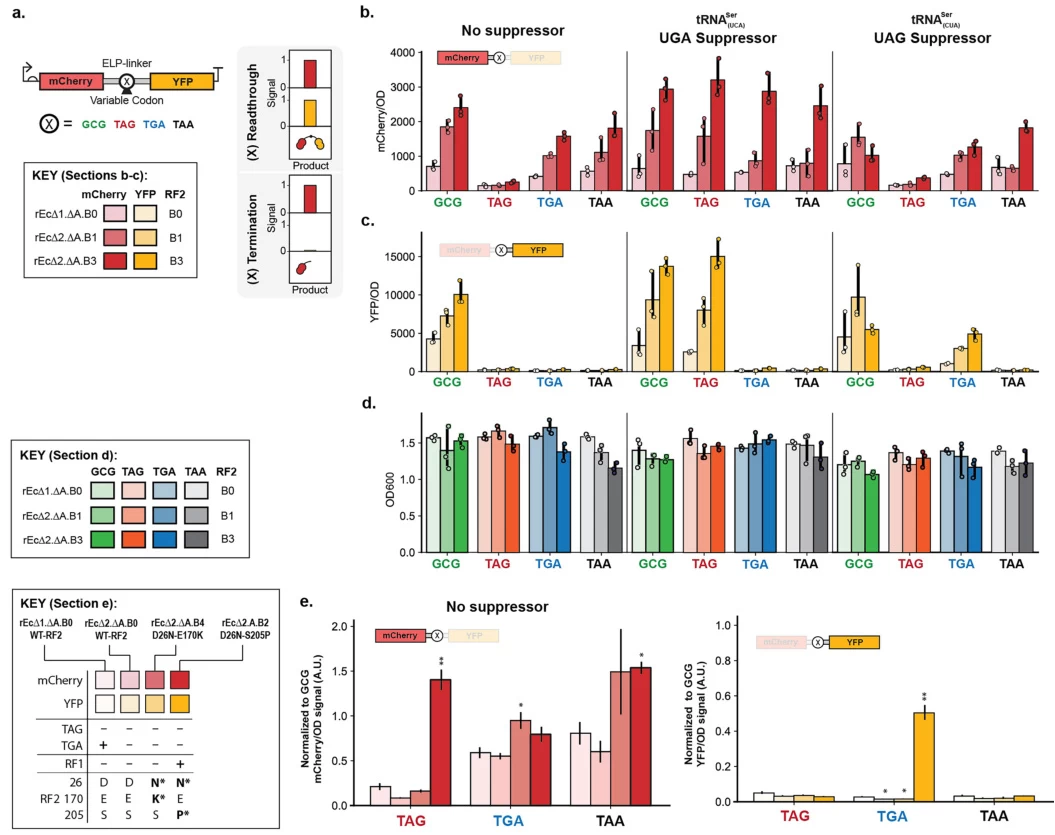
- Synthetic Biology: Engineered tRNAs suppress stop codons to incorporate noncanonical amino acids (e.g., fluorescent tags) .
B. Viral Genome Annotation
Tools like Pharokka-gv predict stop codon reassignment in bacteriophages, improving gene annotation accuracy and revealing novel translational strategies .
Image suggestion: Workflow of stop codon reassignment prediction in viral genomes, highlighting increased coding density.
6. Structural Insights from Cryo-EM and Biochemical Studies
Recent advancements in cryo-electron microscopy (cryo-EM) have elucidated how stop codons and release factors interact within the ribosome:
- eRF1-Stop Codon Interaction: The stop codon and +4 base form a composite signal recognized by eRF1. This interaction flips ribosomal RNA nucleotide A1825, stabilizing the termination complex .
- Ribosomal RNA Role: While tRNA-like molecules do not directly recognize stop codons, ribosomal RNA (rRNA) residues (e.g., G626) facilitate codon-anticodon mimicry during termination .
Image suggestion: Atomic-resolution model of the ribosomal A-site during termination, showing eRF1 (purple), stop codon (UAA), and key rRNA residues (yellow).
7. Evolutionary Perspectives and Future Directions
A. Origin of Stop Codons
Hypotheses suggest stop codons evolved from early “ambiguous” codons in the primordial genetic code, later co-opting release factors for precision .
B. Synthetic Stop Codons
Engineering novel stop codons (e.g., quadruplet codons) could expand the genetic code for synthetic biology applications .
C. AI-Driven Predictions
Machine learning models (e.g., DeepCRISPR) predict stop codon impacts in mutagenesis screens, accelerating therapeutic design .
Summary Table: Key Properties of Stop Codons
| Property | Description |
|---|---|
| RNA Sequences | UAA, UAG, UGA |
| DNA Sequences | TAA, TAG, TGA |
| Release Factors | RF1/RF2 (prokaryotes), eRF1 (eukaryotes) |
| Disease Relevance | Nonsense mutations in CFTR, DMD, and PCSK9 |
| Biotech Applications | Gene knockout, CAR-T engineering, viral genome annotation |
| Evolutionary Flexibility | Dual-coding (UGA → selenocysteine), mitochondrial variants (AGA, AGG) |
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




