Precision in Protein Synthesis and Evolutionary Adaptations
1. The Molecular Architecture of Translation Termination
Stop codons—UAA, UAG, and UGA—are nucleotide triplets in mRNA that signal the termination of protein synthesis. These codons do not encode amino acids but instead recruit release factors (RFs) to halt translation and release the nascent polypeptide chain from the ribosome. The process ensures accurate protein length and functionality, preventing the production of aberrant, potentially harmful polypeptides .
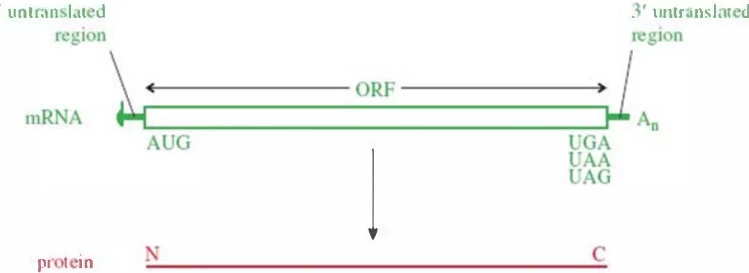
Image suggestion: Ribosomal complex model showing stop codon (UGA) recognition by release factor eRF1, with mRNA and nascent polypeptide chain.
2. Release Factors: Key Players in Termination
A. Prokaryotic Systems
- RF1: Recognizes UAA and UAG.
- RF2: Recognizes UAA and UGA.
- RF3: A GTPase that enhances RF1/RF2 activity and facilitates ribosomal subunit dissociation .
B. Eukaryotic Systems
- eRF1: A universal release factor that recognizes all three stop codons (UAA, UAG, UGA). Structural studies reveal its tRNA-like domain interacts with the ribosomal decoding center .
- eRF3: Binds GTP and cooperates with eRF1 to hydrolyze the peptidyl-tRNA bond, ensuring efficient termination .
Image suggestion: Cryo-EM structure of eRF1 bound to a UAA stop codon, highlighting ribosomal RNA interactions.
3. Stop Codon Diversity Across Species
A. Mitochondrial Variations
- Vertebrates: Repurpose AGA and AGG (typically arginine codons) as additional stop signals, adapting to reduced genome complexity .
- Green Algae: Use UCA as a termination signal while reassigning UGA to encode tryptophan .
B. Ciliate Protozoa
- Condylostoma magnum: All three canonical stop codons (UAA, UAG, UGA) can encode amino acids (e.g., glutamine) depending on mRNA context. Termination relies on transcript termini rather than codon identity .
Image suggestion: Phylogenetic tree highlighting stop codon reassignment in mitochondria, algae, and ciliates.
4. Functional Plasticity of Stop Codons
A. Dual-Functional Codons
- Selenocysteine (Sec): In eukaryotes, UGA encodes Sec when accompanied by a SECIS element (selenocysteine insertion sequence). This rare amino acid is critical for antioxidant enzymes like glutathione peroxidase .
- Readthrough Mechanisms: Certain viruses and engineered systems exploit “leaky” termination, where UAG or UGA encode amino acids (e.g., glutamine) under specific conditions. This enables extended protein synthesis for viral replication or synthetic biology applications .
B. Recoded Genomes
Genomically recoded organisms (GROs) replace natural stop codons with UAA, freeing UAG and UGA for non-canonical amino acid incorporation. This expands the genetic code for synthetic protein engineering .
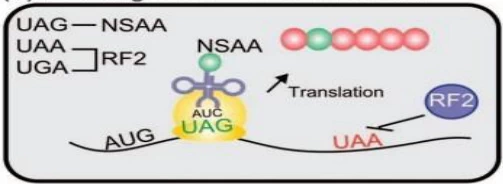
Image suggestion: Schematic of a genomically recoded organism (GRO) with unified UAA stop codons and repurposed UAG/UGA.
5. Clinical and Biotechnological Applications
A. Nonsense Mutation Therapies
Premature stop codons (PTCs) truncate proteins, causing diseases like cystic fibrosis (CFTR mutations) and Duchenne muscular dystrophy (DMD mutations). Readthrough drugs (e.g., ataluren) promote ribosomal bypass of PTCs, restoring partial protein function .
B. CRISPR-Cas9 Gene Editing
- Gene Knockout: Introducing stop codons into CDS disrupts target genes (e.g., PD-1 knockout in CAR-T cells for cancer immunotherapy) .
- Exon Skipping: sgRNAs targeting splice sites exclude exons containing PTCs, enabling functional protein production in muscular dystrophy models .
Image suggestion: Workflow of CRISPR-Cas9 introducing a stop codon to disrupt PD-1 in T cells.
6. Evolutionary Insights and Future Directions
A. Origin of Stop Codons
Stop codons likely evolved from ambiguous primordial codons co-opted for termination. Their redundancy (three codons for one function) buffers against mutations, while species-specific reassignments exemplify genetic code plasticity .
B. Synthetic Biology Frontiers
- Quadruplet Codons: Engineering non-canonical stop signals (e.g., four-nucleotide codons) could expand genetic code flexibility .
- AI-Driven Prediction: Machine learning models predict stop codon impacts on CDS integrity, improving genome annotation in non-model organisms .
Summary Table: Stop Codon Properties and Functions
| Codon (RNA) | Termination Efficiency | Dual-Functional Roles | Species-Specific Adaptations |
|---|---|---|---|
| UAA | High (universal) | None | Primary stop signal in prokaryotes |
| UAG | Moderate | Glutamine (ciliates, viruses) | Amber suppression in synthetic biology |
| UGA | Variable | Selenocysteine (eukaryotes) | Tryptophan (mitochondria, green algae) |
Data Source: Publicly available references.
Contact: chuanchuan810@gmail.com




